Recolorize: Color-based image segmentation (for people with other things to do)
Abstract
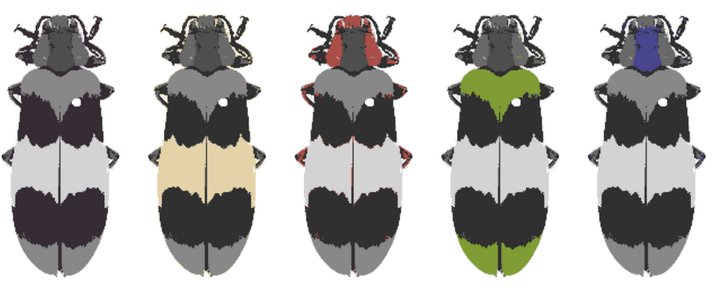
Color and pattern are some of the most conspicuous signals in biology, whether they are naturally present or introduced (e.g., by staining); often, these are the first-noticed and best-documented traits of organisms. An increasing number of methods attempt to quantify some aspect of these signals, typically focusing on a particular question, such as sensory ecology (how other organisms view a signal), comparative biology (how the signal has evolved), or anatomy. The first step in most of these methods is to cluster an image into a small number of color classes. This step has proven to be a surprising bottleneck, partly because the problem is not generalizable—image noise in one context could be relevant signal in another. A green-blue gradient might indicate inconsistent lighting on a lizard, while the same gradient on a beetle indicates biologically important iridescence. A common solution to this problem is k-means clustering, which has three major issues: it produces inconsistent results, requires a user-specified number of clusters, and often loses minor elements while over-clustering large ones. More robust alternatives (e.g., micaToolbox) are tailored for sensory biology and require additional information which may not be available. To address this bottleneck, we created recolorize, an R package which provides automatic and manual tools for color classification in images or image sets. Options include hierarchical clustering, layer merging and cleaning, and color fit optimization, along with export tools for popular color analysis packages.